Why Ordaōs
Introducing miniPRO™
The Ordaōs solution, miniPRO™, is a class of mini-proteins that promise to provide the power and performance of antibodies, but are more configurable, stable, and easier to manufacture.
Introducing the Ordaōs Platform
We use our multitask meta-learning Design Engine to create novel mini-proteins from scratch.
The Design Engine leverages generative Al, proprietary multitask meta-learning, and reinforcement learning to create and evaluate de novo protein sequence, structure and physiochemical properties in silico to reliably and repeatedly deliver de novo miniPRO™ that meet the specifications of the target product profile.
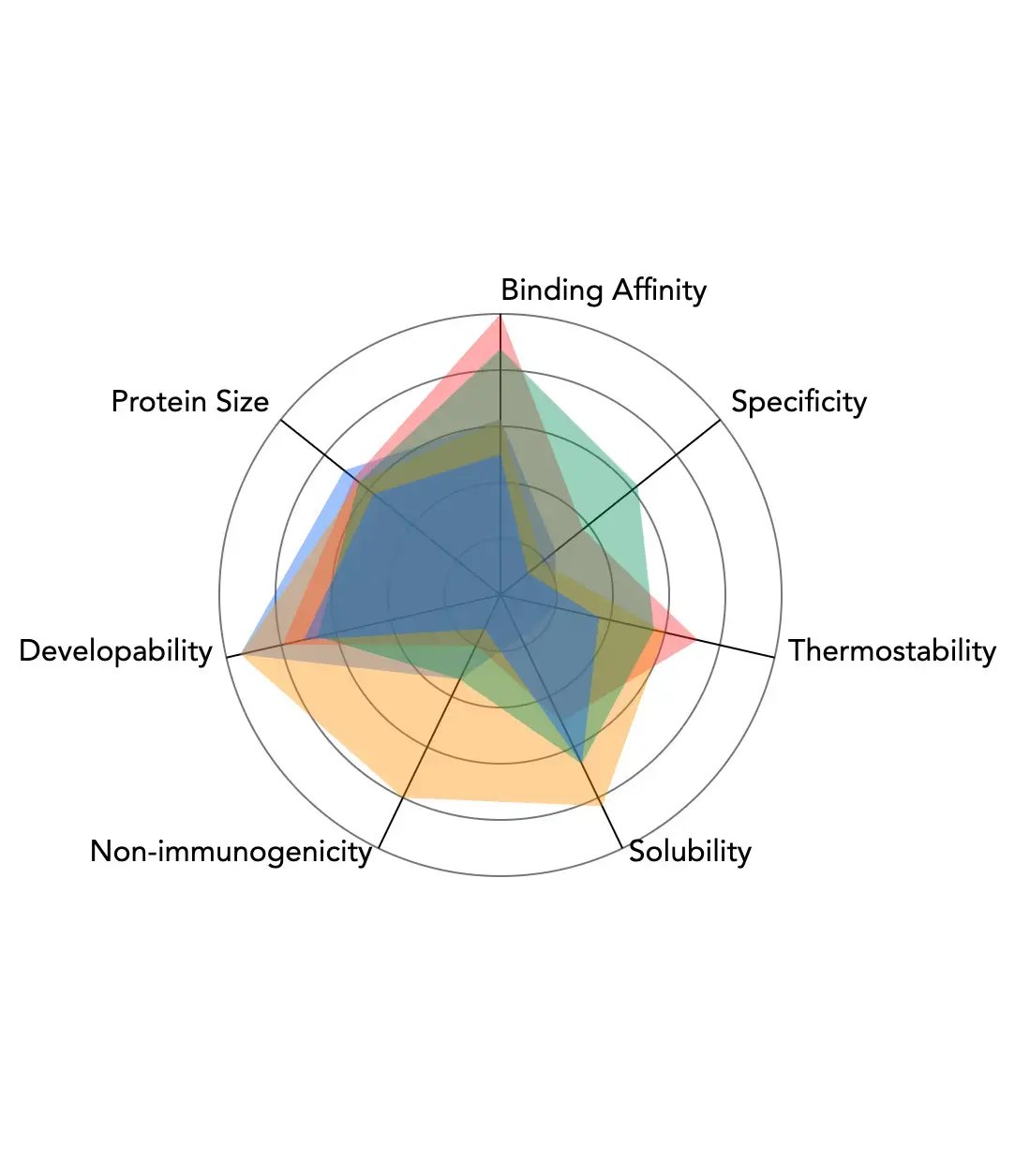
De Novo design case study
Designing de novo miniPRO™ binders to HER2 with generative AI resulting in leads that are 93% smaller than standard of care, within weeks
.webp?width=1869&height=1171&name=image%20(1).webp)
About miniPRO Proteins
Introducing miniPRO™, a new class of mini-proteins that will revolutionize the role of proteins in drug discovery. Helping drug hunters create novel therapies for areas of high unmet need.
- Configurable building block
- 20x smaller than traditional monoclonal antibodies (40-160 amino acids) enabling them to more easily penetrate disease microenvironments for effective treatment
- Sound structure with improved thermostability and solubility, and minimized aggregation and other developability risks to ensure the ease of manufacturing
- Configurable binding, optimized for on-target vs off-target binding for selected epitope, with tuned affinity range and configurable isoelectric point
- Humanized and optimized to meet client-specific MHC / peptide profiles to minimize immunogenicity
.webp?width=587&height=312&name=image%20(2).webp)
Optimization case study
Optimizing single domain VHH antibodies against SARS-CoV-2 S1 with generative AI resulting in therapeutic candidates that overcome evolutionary escape.
.webp?width=927&height=1236&name=image%20(3).webp)
About Ordaōs Design Engine
The Ordaōs Design Engine is a multitask meta-learning model that leverages continuous learning loops and both public and proprietary data sets to translate human-defined product criteria into machine-designed miniPRO™.
Starting with amino acids, the engine generates, appraises, and ranks billions of protein sequences, hundreds of thousands of protein structures and other drug-like properties to create miniPRO™.
miniPRO™ are evaluated to provide intelligent feedback on multiple design objectives including protein structure, binding specificity and affinity, solubility, stability, immunogenicity, and developability. The analysis feeds the engine within a generative AI reinforcement learning loop, to iteratively improve and deliver optimal miniPRO™ that meet the client’s molecular target product profile (mTPP).
.webp?width=470&height=329&name=image%20(4).webp)
Benefits of Ordaōs System
.webp?width=166&height=166&name=image%20(5).webp)
Scalable
.webp?width=166&height=166&name=image%20(6).webp)
Repeatable
.webp?width=166&height=166&name=image%20(7).webp)
Highly Automated
.webp?width=166&height=166&name=image%20(8).webp)
Disease Agnostic
.webp?width=166&height=165&name=image%20(9).webp)
Perceptive System
.webp?width=166&height=165&name=image%20(10).webp)
Efficacy
.webp?width=166&height=165&name=image%20(11).webp)
Time to Value
.webp?width=166&height=165&name=image%20(13).webp)
IP Protected
"You accomplished in four-days what took us two-years to accomplish."
Dr. Philip Howe, Chair, Department of Chemistry,
Medical University of South Carolina
Partner with us
By maximizing drug candidate delivery speed, novelty, and probability of clinical success, we can provide the highest client confidence in every investigational new drug (IND) application.
What’s more, our clients see a return on their investment in weeks, rather than months or years.
.webp?width=3024&height=3006&name=image%20(14).webp)
"The AI-based drug design engine Ordaōs has developed is a method with game-changing potential."
Dr. Ülo Palm, Chief Medical Officer, Vaxxinity
.webp?width=271&height=136&name=image%20(15).webp)
.webp?width=271&height=86&name=image%20(16).webp)
.webp?width=271&height=111&name=image%20(17).webp)
.webp?width=271&height=218&name=image%20(18).webp)
.webp?width=271&height=328&name=image%20(19).webp)
.webp?width=271&height=106&name=image%20(20).webp)
.webp?width=271&height=105&name=image%20(21).webp)
.webp?width=271&height=125&name=image%20(22).webp)
Ordaos is a resident company of Johnson & Johnson Innovation – JLABS
Ordaōs in the News
We are in the news because we’re making huge strides in human-enabled, machine-driven drug design.
Posters
Read abstracts and sign up to download more info
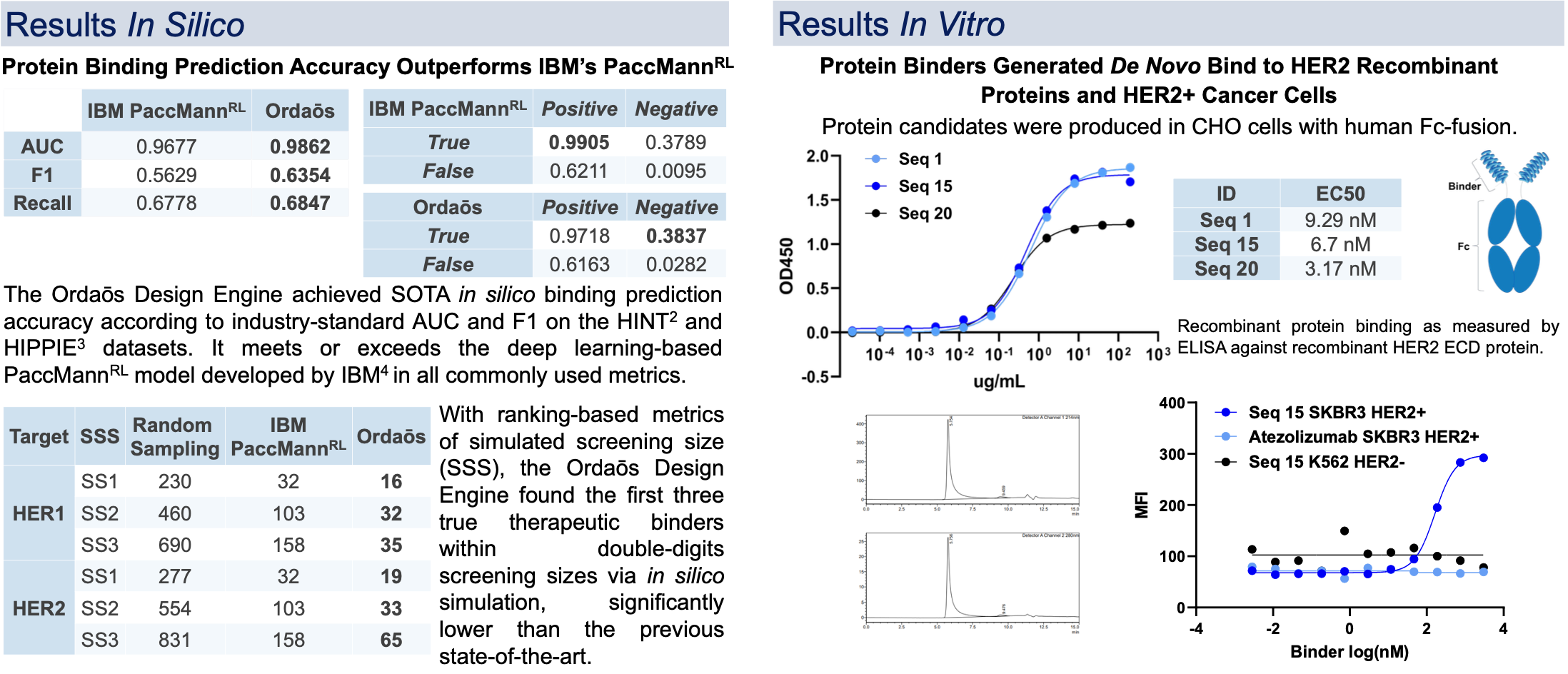
The Ordaos Design Engine, a deep learning-based end-to-end protein therapeutics design platform, was developed and used to create de novo mini-proteins to target HER2. Candidate proteins are evaluated in silico for binding affinity and specificity, secondary and tertiary structure, isoelectric point, solubility, and other drug-like properties, and iteratively improved to achieve multi-objective optimization. These in silico designed binders can be produced with high yield and purity using mammalian protein expression systems, and have been shown to bind to human HER2 with EC50 in the nanomolar range by ELISA.
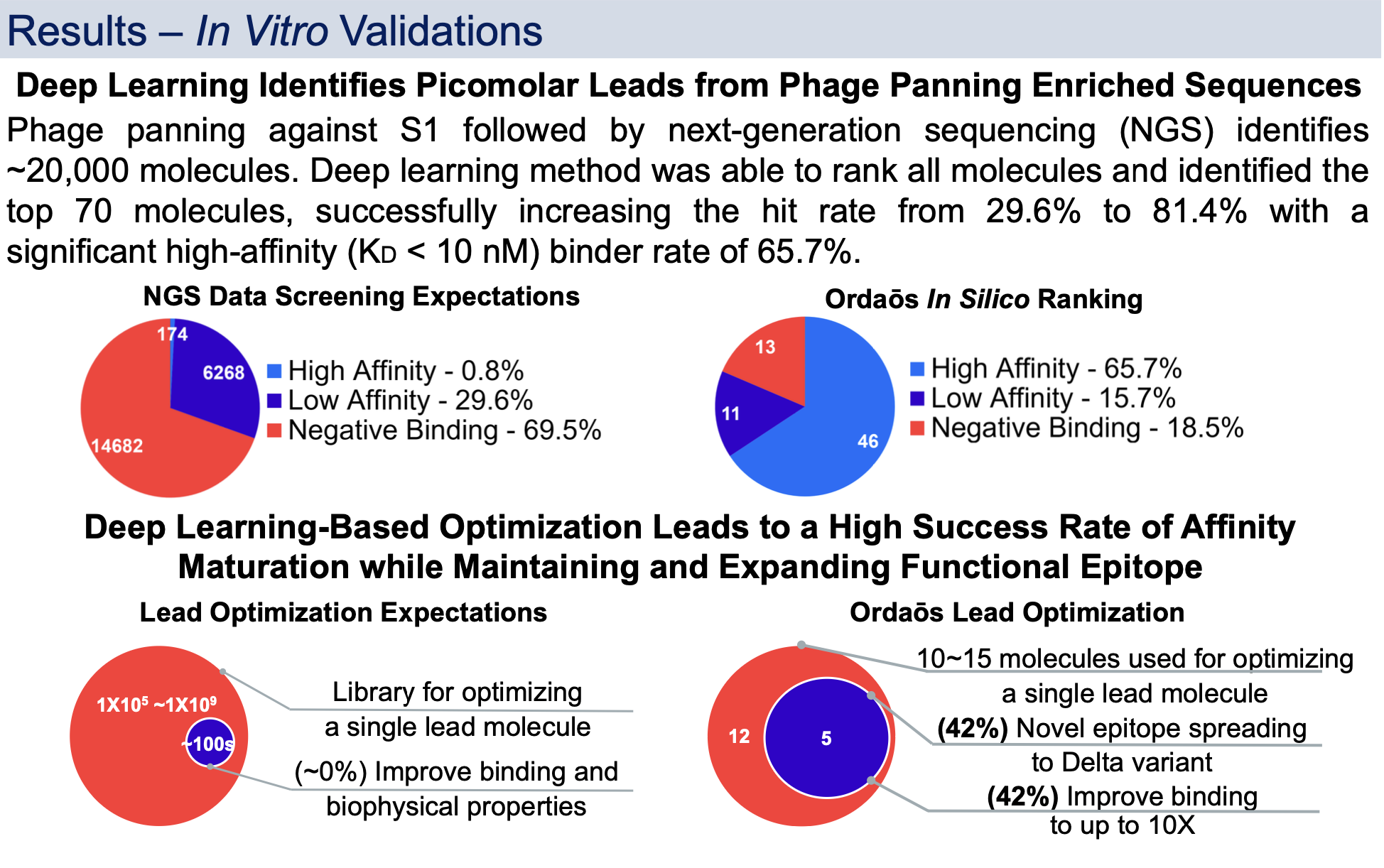
After presenting at AET 2022 with updated in vitro results we share this exciting data.
Therapeutic SARS-CoV-2 antibody is desired to target S1 whilst addressing its viral evolution and escape mutations. A deep learning-based approach was developed to create, optimize, and rank mini-protein binders for drug-like properties including binding affinity and developability. The approach was used to discover single domain VHH antibodies that bind to S1 Ancestral, Delta and Omicron simultaneously with picomolar to sub-nanomolar affinity and block S1 interaction with human ACE2 in vitro.
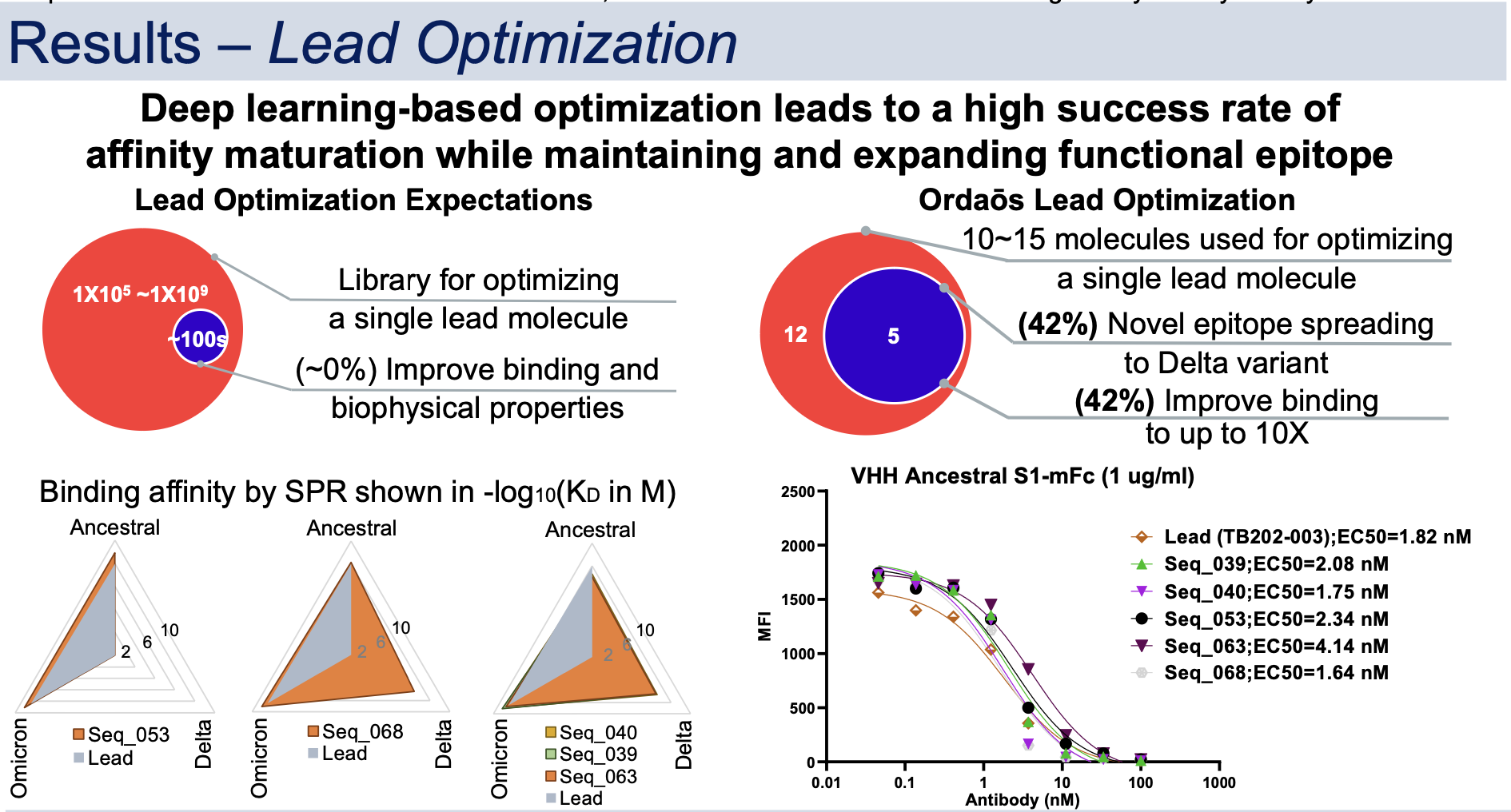
Therapeutic SARS-CoV-2 antibody is desired to target S1 whilst addressing its viral evolution and escape mutations. A deep learning-based approach was developed to create, optimize, and rank mini-protein binders for drug-like properties including binding affinity and developability. The approach was used to discover single domain VHH antibodies that bind to S1 Ancestral, Delta and Omicron simultaneously with picomolar to sub-nanomolar affinity and block S1 interaction with human ACE2 in vitro.

